It’s the year 2009, and I’m wondering: where is my flying car? After all, Hollywood reels from the 60’s and 70’s all predicted that flying cars are what I’d be using to get around town these days. Of course, automotive technology isn’t the only victim of Hollywood hype. The potential impact of personalized genomics has been greatly overstated in movies like GATTACA. This has lead to the pervasive myth that your genome is like a crystal ball, and somehow your fate is predestined by your genetic programming. Recently, my perlfriend co-authored a paper in Nature (“A Personalized Medicine Research Agenda”, Nature Vol 461, October 8 2009), comparing Navigenics’ and 23andMe’s “Direct to Consumer” (DTC) personal genomics offerings. She’s qualified to offer deep insight into personal genomics, since she designed the original Illumina bead chip used by leading companies to generate their DTC genetic data, and she is also the person who made sense of the first complete diploid human genome sequence (1 2). She’s sort of the biology equivalent of the reverse engineer who takes binary sequences and annotates meaning into the disassembled binary sequences. So, let the mythbusting begin.
Myth: having your genome read is like hex-dumping the ROM of your computer. Many people (I was one of them) have the impression that “reading your genome” means that at the end of the day someone has a record of all the base pairs of DNA in my genome. This is called a “full sequence”. In reality, full sequencing is still cost-prohibitive, and instead a technique called “genotyping” is used. Here, a selective diff is done between your genome and a “reference” human genome, or in other words, your genome is simply sampled in potentially interesting spots for single-point mutations called Single Nucleotide Polymorphisms (SNPs, pronounced “snips”). In the end, about 1 in 3000 base pairs are actually sampled in this process. Thus, the result of a personalized genomic screen is not your entire sequence, but a subset of potentially interesting mutations compared against a reference genome. This naturally leads to two questions: first, how do you choose the “interesting subset” of SNPs to sample? And second, how do we know the reference genome is an accurate comparison point? This sets us up to bust another two myths.
Myth: We know which mutations predict disease. Herein lies a subtle point. Many of the mutations are simply correlative with disease, but not proven to be predictive or causal with disease. The truth is that we really don’t understand why many genetic diseases happen. For poorly understood diseases (which is still most of them), all we can say is that people who have a particular disease tend to have a certain pattern of SNP mutations. It’s important not to confuse causality with correlation. Doing so might lead you to conclude, for example, that diet coke makes you fat, because diet coke is often consumed by people who are overweight.
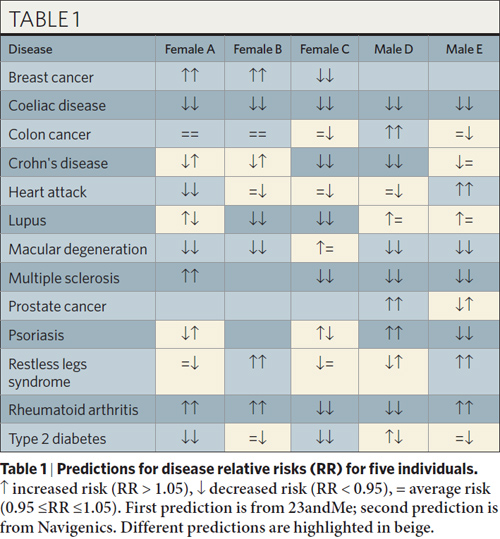
Thus, there are two echelons of understanding that can come from a genotype: disease correlations, and disease causes. The majority of SNP mutation-based “predictions” are correlative, not causative. As a result, a genotype should not be considered a “crystal ball” for predicting your disease future; rather, it is closer to a “Rorschach blot” that we have to squint and stare at for a while before we can make a statement about what it means. The table below from the paper illustrates how varied disease predictions can be as a result of these disagreements on the interpretation of mutation meanings.
Myth: the “reference genome” is accurate reference. The term “reference genome” alone should tip you off on a problem: it implies there is such a thing as “reference people”. Ultimately, just a handful of individuals were sequenced to create today’s reference genome, and most of them are of European ancestry. As time goes on and more full sequence genetic data is collected, the reference genome wlll be merged and massaged to present a more accurate picture of the overall human race, but for now it’s important to remember that a genotype study is a diff against a source repository of questionable universal validity, partially because it’s questionable if there is such a thing as a “reference human”, i.e. there are structural variations and some SNPs have different frequencies in different populations (e.g. the base “A” could dominate in a European population, but at that same position, the base “G” could dominate in an African population). It’s also important to keep in mind that the “reference genome” has an aggregate error rate of about 1 error every 10,000 base pairs, although to be fair the process of discovering a disease variant usually cleans up any errors in the reference genome for the relevant sequence regions.
So now you can see that in fact “reading your genome” is less of looking into a crystal ball and more of staring at a Rorschach blot obscured by cheesecloth (i.e., the genome is simply sampled and not sequenced). And, even if we could remove the cheesecloth and sequence the genome such that we knew every base pair, it would still be … a Rorschach blot, but in high resolution. It will be decades until we have a full understanding of what all the sequences mean, and even then it’s unclear if they are truly predictive.
Here lies perhaps the most important message, and a point I cannot make fine enough: in most situations, environment has as much, perhaps even more, to do with whom you are, what you become, and what diseases you may develop than your genes. If there is any upside to personal genomics, it won’t be due to crystal ball predictions. It will be the lifestyle changes it can encourage. If there’s one thing I’ve learned from dating a preeminent bioinformaticist, it’s that no matter your genetic makeup, most common diseases can be prevented with proper diet and exercise.
http://ds9a.nl/amazing-dna/ is a wonderful page documenting how DNA behaves from the point of view of a programmer. It makes Intercal look sane and no one would ever design a “computer” and “language” that way.
Excellent, I enjoyed this almost as much as ‘ On Influenza A (H1N1) – http://www.bunniestudios.com/blog/?p=353 ‘.
Fortunately this was a much shorter read.
I don’t have a disinterest in biology, but I don’t encounter it often / seek it out. Combine that with Comp/Elec Engineering comparisons, and it increases in both understanding and interest! If at the price of a smaller audience? perhaps not smaller, just a different one altogether.
Anyways, thanks. –PidGin128
[Very unrelated, except as an example of my scattered exposure to Bio. But Plants can communicate/signal to warn each other, this I found interesting. ‘ http://en.wikipedia.org/wiki/Plant_communication#Non-human_living_organisms_communication ‘ ]
I’m still waiting for the Eugenics Wars and Khan’s rampage of terror to happen. He’s late!
Anyhoo, I noticed one flaw in the table showing risks for the five people. There should be male entries for breast cancer. And shouldn’t female B have a risk listed for psoriasis as well?
But on point, I’m in agreement here. DNA doesn’t define what will and will not happen to us. It may point to possibilities, but until we have a greater understanding of DNA and have made advances in science, it’s had to swallow the idea that what makes us human also dictates how we’ll live our lives or meet our demise. I doubt I’ll live long enough to see the day when (if) doctors can say with certainty that someone will get a certain disease or live a guaranteed x amount of years (providing no accidental death, murder, etc.).
Seriously, I don’t think that’ll ever happen, but hopefully things will advance to the point where doctors and scientists can better DNA sequences to better predict and treat patients. Hopefully things don’t get to the point as depicted in Gattaca, where your DNA can be used to discriminate against you.
23andMe and Navigenics don’t currently list breast cancer predictions for males. I’m guessing it’s because all of the risk markers identified so far have been in studies involving females only, so it’s not known whether the risk markers would transfer to a male population.
Subject B does not have a risk listed for psoriasis because not all her genotypes were called. This happens when the sample quality is poor (i.e. not enough spit), and the companies err on the side of caution and don’t give predictions if the DNA isn’t genotyped correctly.
GINA is the Genetic Information Discrimination Act which states that employers and insurance companies cannot use DNA to discriminate against you.
Thanks for the clarification. I wasn’t aware of GINA’s existence, or that it was even needed yet. But I suppose in the age of the lawyer…
Still, I have to wonder how much legal protection is actually going to do as genetics progresses; just as discrimination laws don’t curtail everything today, DNA laws may not cover everything in the future. But being pessimistic may not be the best position to take.
Moving on, the incomplete table does make sense. There’s only so much you can do with a limited set of samples at this point. Science (any field) was never my best subject (but far from my worst), so I should’ve thought a bit more about the data presented. Oh well, such is life sometimes.
[…] google shared Mythbusting Personalized Genomics. […]
heh heh. “perlfriend” ;-)
(Before people snark back at me, I know bunnie personally, and I’d definitely say this to his face :-)
Perlfriend?
It’s someone who can write you this:
$_ = “girlfriend”; s/gi/pe/;
…and also cuddle up with at night!
I get your point, but I had to point out a bad example.
Diet soda does make you fat. Or, at least, it’s been correlated with increased risks along those lines.
http://circ.ahajournals.org/cgi/content/short/117/6/754
It might be all hype and overkill now, but discussing the possibilities (and ethical consequences) before these things are possible is really not such a bad idea.
Ok, I can’t lie, much of what I read is over my head. But I get the idea and enjoyed the read since having my genome peaks my curiosity as an adopted person.
In the end, I got a kick out of the fact that my dietitian mother at 73 has been saying the same thing as this article for the past 50 years….” that no matter your genetic makeup, most common diseases can be prevented with proper diet and exercise.”
Mom, your right! I’d better go eat that apple…
[…] http://www.bunniestudios.com/blog/?p=563 a few seconds ago from xmpp […]
[…] Mythbusting personal genomics. […]
Your flying car is now called a ‘bicicle’. Oh, you live in the USA :) Tough luck!
Hi hunnie, great website! I really appreciate this blog post.. I was curious about this for a long time now. This cleared a lot up for me! Do you have a rss feed that I can add?
[…] Så när @peterkz_swe lattjar lite med mina SNPs i Promethease så påminner @ehn oss om den texte: http://www.bunniestudios.com/blog/?p=563 Bra […]
http://online.wsj.com/article/SB10001424052748703559004575256470152341984.html
Looks like impressive genome hacking to me. I was wondering if you have any comments.
Just read the news that a women who smoke or have a history of smoking are at 39 percent higher risk of death due to breast cancer.
Nice blog. Thanks! i will be back here soon.